Structural basis of bacterial transcription repression
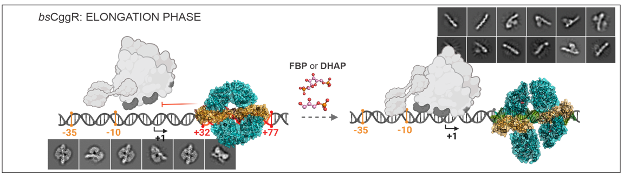
Bacterial repressors play a crucial role in gene transcription regulation. The GntR and SorC/DeoR families are major groups of metabolic gene regulators. Despite their significance, the molecular mechanisms underlying their regulatory functions remain largely unknown. Understanding their three-dimensional structure is essential for unraveling these processes and can have implications for other family members. We focused on members of the SorC/DeoR family in B. subtilis: bsDeoR and bsCggR, representing distinct subfamilies. Through crystal structure determination, we uncovered the DNA recognition mechanism of these regulators by examining their N-terminal DNA binding domains in complex with minimal operator sequences. Moreover, we structurally characterized the full-length proteins bound to complete DNA operators. Our findings revealed a shared global architecture and a common DNA binding mechanism between the two repressors. This study presents a fundamental mechanism for the function of SorC family transcriptional regulators, laying the groundwork for future basic and applied research in this field.
Contact: Adéla Fejfarová, Jana Škerlová, Pavlína Maloy Řezáčová