Protein-protein interactions in eukaryotic transcription
The goal of the project is to investigate interactions involved in eukaryotic transcription, with a particular focus on understanding the role of the lens epithelium-derived growth factor (LEDGF) in gene transcription regulation. We uncovered the molecular mechanisms underlying LEDGF's interactions with other transcriptional regulatory factors and explored its involvement in a protein regulatory network controlling transcription elongation.
Contact: Vanda Lux, Eliška Koutná, Lisa-Maria Weinhold, Terézia Paulovčáková, Václav Veverka
Multivalency of nucleosome recognition by LEDGF
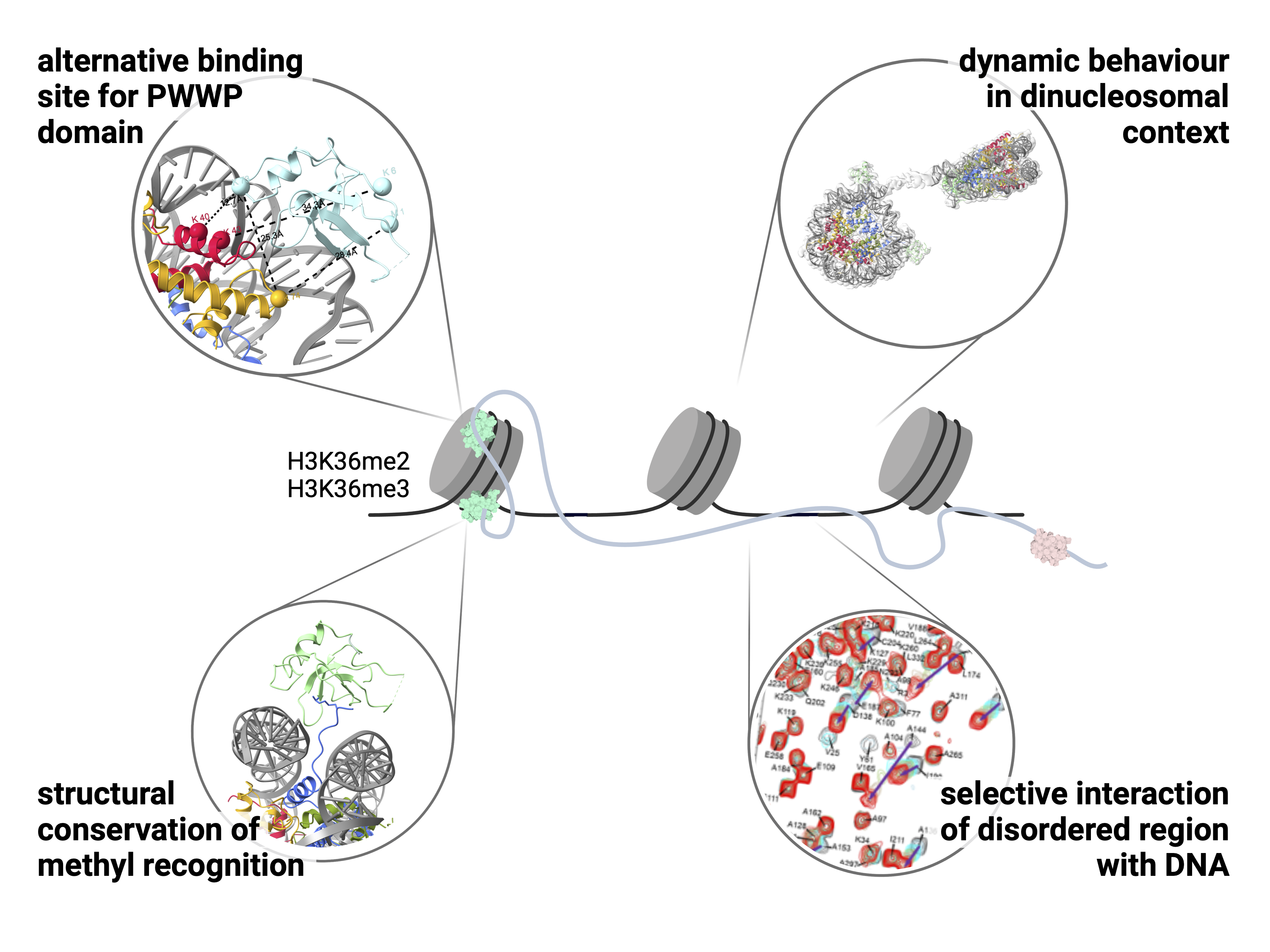
Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation
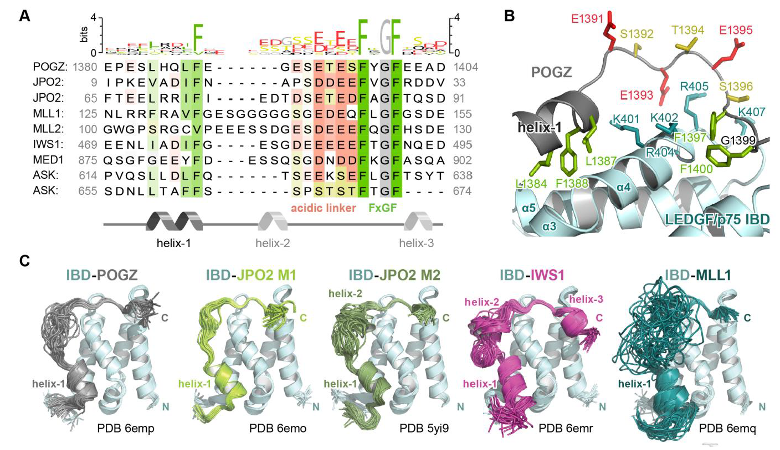
"Mixed lineage" leukemia poses a significant challenge with a low survival rate of 50% in affected children. We identified a specific vulnerability in this type of leukemia by analyzing the LEDGF protein's structure. We discovered essential molecular features governing LEDGF's interactions with binding partners, revealing previously unknown direct interactions with major transcriptional regulatory factors. This expands our understanding of the broader network involved in transcription elongation. Importantly, we found that phosphorylation, mediated by casein kinase 2, strongly influences the binding between LEDGF and its partners. We demonstrated that eliminating phosphorylation sites in MLL1, an LEDGF interaction partner, reduces the cancerous state of leukemic cells without impacting normal blood cell function.
Molecular Mechanism of LEDGF/p75 Dimerization
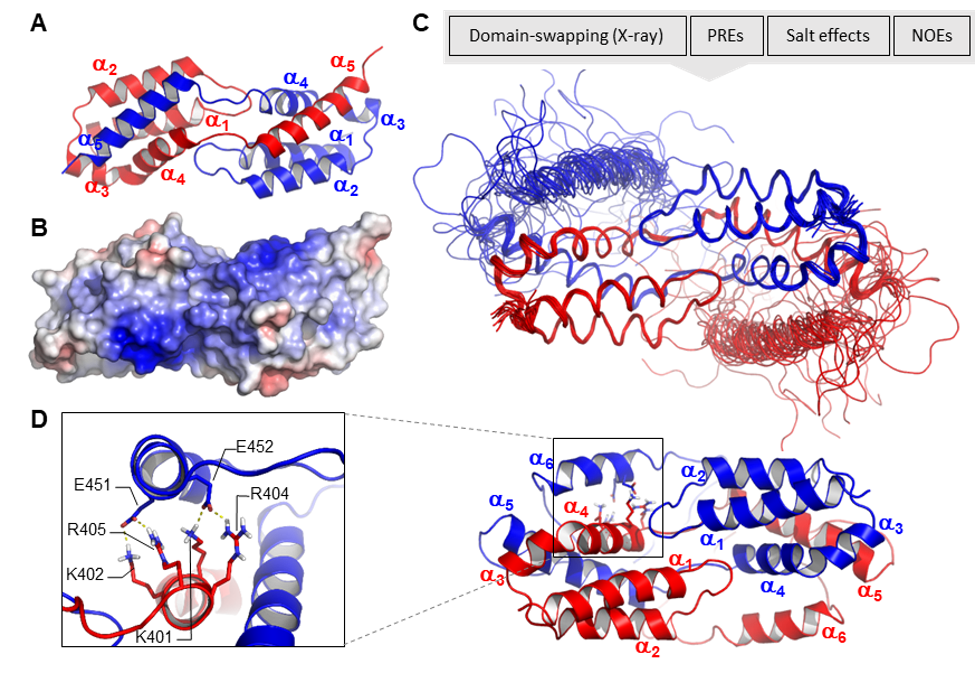
LEDGF plays a crucial role in tethering transcriptional regulators to chromatin in both normal and pathological contexts. Previous studies suggested that LEDGF dimerization is important for its function as an epigenetic reader. However, direct evidence of LEDGF dimerization was lacking. In our study, we aimed to answer three key questions: (1) Can LEDGF/p75 form dimers? (2) What is the minimal domain responsible for dimerization? (3) What is the molecular mechanism of dimerization? Using NMR spectroscopy, X-ray crystallography, and various biochemical techniques, we uncovered the molecular basis of LEDGF dimer formation. The minimal dimerization domain consists of the integrase binding domain and a dynamic C-terminal region containing an additional helix that stabilizes the dimer through electrostatic interactions. Dimerization does not affect LEDGF's ability to interact with cellular partners in vitro and may serve to locally increase the concentration of interacting factors, enhancing active transcription. We propose that LEDGF dimers act as molecular "glue," cross-linking the same or different interaction partners and amplifying epigenetic signals.
A ubiquitous disordered protein interaction module orchestrates transcription elongation
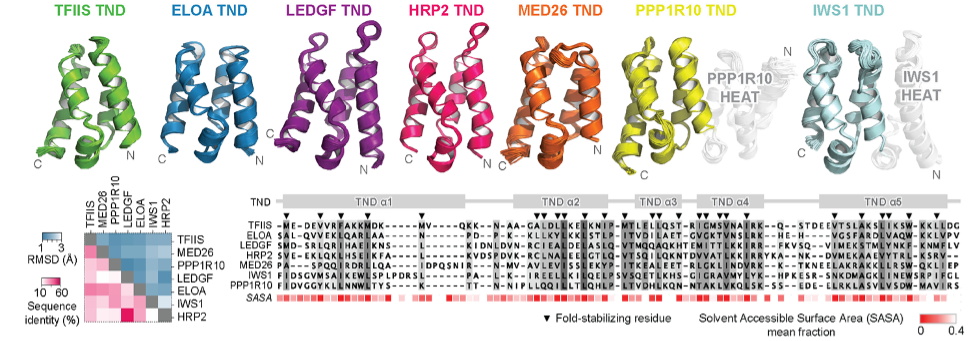
Unstructured regions in protein sequences, previously believed to lack function, have been found to play important biological roles. While their significance in protein signaling, localization, and stability is well-established, their roles in other contexts have remained elusive. We discovered that the transcription elongation machinery involves interactions between transcription elongation factor TFIIS N-terminal domains (TNDs) and conserved unstructured sequences called "TND-interacting motifs" (TIMs). By mutating a single TIM in a central organizing protein of this network, we disrupted key protein interactions and observed widespread defects in transcription elongation dynamics. The conservation and utilization of TNDs highlight their critical role in assembling the transcriptional machinery. Understanding the regulation of individual TND interactomes represents a new research direction. Additionally, our work contributes to the deciphering interactions between disordered sequences and folded protein domains.

